Getting to the heart of thickfilament structure
A large-angle X-ray diffraction study of the lattice of intact muscle. III. The cardiac thick filament reconstruction at the three crowns in the 430 repeat
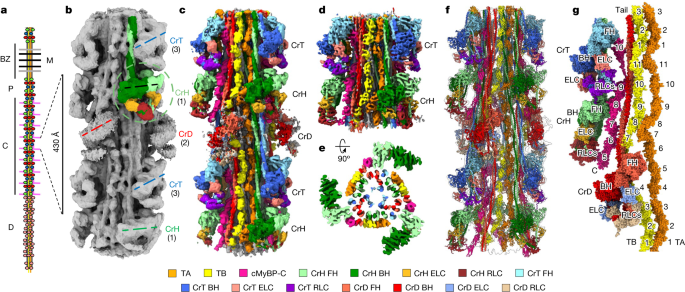
The x-ray data gave information on the structure of the lattice of intact muscle. A, Longitudinal and b-d views of our cardiac thick filament reconstruction at the three crowns in the 430 repeat, with a model based on X-ray diffraction. The radial positioning of the heads, especially in the centre-of-mass 135 range, suggests that the reconstruction is very close to the structure in intact muscle. The previous negative stain reconstructions had suggested a head centre-of-mass at ~95 Å radius (~40 Å less than the cryo-reconstruction), thought to be due to radial collapse of heads during drying of negative stain22—not an issue with frozen-hydrated specimens. The averaged power spectrum of the reconstruction was compared with the averaged power spectrum of selected segments used in the reconstruction. The similarity of the two power spectra, both extending to the 36th order of the 430 Å repeat (12 Å), supports the validity of the reconstruction. The wide-angle X-ray diffraction pattern of intact, relaxed porcine cardiac muscle is courtesy of the Drs. Tom Irving and Weikang Ma, unpublished data) shows similar myosin layer lines at low angles, further suggesting that our structure is similar to that in intact muscle. The 39 reflection is a key feature of all patterns. This may be due to the fact that there’s more than one culprit. Our reconstruction reveals titin unambiguously (see text and Extended Data Fig. 5), and we show that the reflection arises from the kinking of its elongated structure, allowing 11 domains to fit into the 430 Å repeat (Extended Data Fig. 6c). h, i. We looked at the cryo-EM structure of a local IHM with 15 heptads of tail. 8ACT (yellow) was superimposed on CrH (green) and CrT (blue) (h, i, respectively). Each case the FH was matched with BHs and revealed how it is positioned with respect to the BH. There was strong similarity between the 3 different Ihms. There are different interactions with c MyBP-C and the backbone of the native Filament that causes the two IHMs to be non-identical. 6g–m. 8ACT was not perfect, but its individual blocked and free heads showed excellent correspondence to the respective head. Across 884 pairs, the crH BH and 8ACT BH RMSD was between 2.148 and 1.268, and between 592 pruned atom pairs it was between 1.268 and 1.189. We think that 8ACT is an excellent model for the two filament Ihms, because of its individual heads matching perfectly with the individual free and blocked heads of the others and the whole IHM being closer to the other ones. The absence of the interactions of the FH and BH that occur with cMyBP-C in the native filament is likely to explain the small difference between the isolated Ihm and its counterparts. RMSD of Cα was calculated with the ChimeraX Matchmaker command using FH and BH separately.

