There is a case for standardizing genes in the vertebrates
Synteny-based Gene Nomenclature: Recommendations for Genome-scale Inference and Implications for Gene and Paralogies
Our analysis shows that integration of genomic data from a wide range of plants can help us update and improve the already-established name with only minor modifications. Theofanopoulou calls for collaboration between the gene nomenclature committee and the genome initiatives. We are encouraging researchers to work with the gene nomenclature committees to make sure the updates are good.
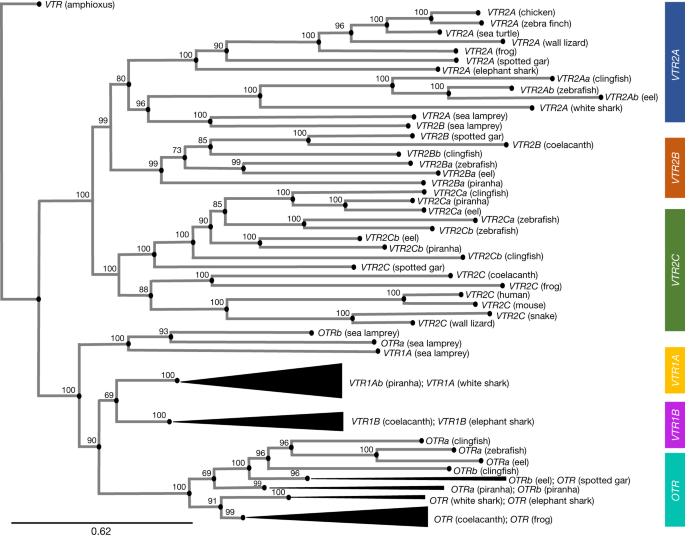
It has been found that synteny-based approaches give the best resolution for genes and paralogies. Wherever available, we propose using chromosome-scale genomes that are highly contiguous and have a high base-call accuracy2. When synteny is not clear, we suggest that priority is given to nucleotide phylogenetic inference with the same prerequisites for genome quality. In Extended Data Fig. 1 and Supplementary Note 5, we provide specific suggestions and caveats with regard to our recommended practices for synteny and phylogenetic analyses. We believe that a combination of synteny and well-supported thgis is the basis of a universal gene nomenclature.
One possible principle is to create one committee for every major lineage (for example, cyclostomes), group these in subcommittees under one larger committee and then group all of them under a committee for one of the animal kingdoms. To achieve such a goal, there would need to be changes to infrastructure and systems, for example, high-quality genomes and synteny.

