Data from the epigenetics is studied to uncover interactions between genes
A Clustered Statistical Study of the K562 Pseudobulk Receptor Cell Types Using ScATAC-Seq Data
To determine the score for each region, we used the scATAC-seq count table and cell annotations from each study. To aggregate single cells into ‘pseudobulk cell types’, we used the Louvain clustering results from each study. Log count per million (logCPM) was used for each pseudobulk. The biological cell type is determined by the annotations of each cluster. Empirically, we set a threshold of more than 600 cell counts to ensure adequate sequencing depth for selected cell clusters. There is a table of pseudobulk cell types found in Supplementary Table 1.
We began gathering inference samples by producing 200 area windows for everyGene’s promoter. The expression value can be inferred from an input matrix with a GET model applied to it.
We stratified the K562 lentiMPRA elements (approximately 200,000) by overlapping the annotated 15 ENCODE ChromHMM states computed from histone mark and other ChIP–seq data for K562. The elements overlap with a number of states, and they were chosen as enhancers and repressive.
Gene-by-motif analysis using a binary ATAC model: A benchmark dataset for calculating Jacobian scores for cell-type-specific genes
The Jacobian matrix looks at how each input dimensions will change when they are factored in. We specifically pick the output dimension and strand that correspond to the given gene, represented by ({\rm{\nabla }}g\in {{\mathbb{R}}}^{r\times m}):
The feature (motif) importance vector ({{\rm{v}}}_{g}\in {{\mathbb{R}}}^{m}) is obtained by multiplying the gradient element-wise with the original input and summarizing across regions:
The Hadamard product is depicted by where (odot ) means element-wise. As the gene-by-motif matrix is mostly used for feature–feature interaction analysis, we use the (X) with quantitative ATAC signal even when we infer ({J}_{X}) using a binary ATAC model. This allows for study of the relationship between regulators and accessibility.
The cell-type-specific genome-wide gene-by-motif matrix for cell type c, Vc is acquired by concatenating the ({{\rm{v}}}_{g}) across the genome. The same process can be used to different types of cells.
We restricted the benchmark dataset to either the fetal erythroblast peak set or K562 DNase peak set for a fair comparison. We used the l2s-norm of the region’s embedded layer Jacobian and the a CPM of the region to get the Jacobian score for each gene. The procedure could possibly be improved in the future by taking a random genomic background and using it as the baseline for Jacobian calculation and other interpretation methods. However, we believe the current benchmark dataset size for this task is still limiting compared with the genome scale (104 measured pairs versus 106 to 107 required measurements of genome-wide enhancer–promoter interaction). We leave the systematic work of this task to the future. The scores used for this study were the same as the others.
Using the gene-by-motif matrix ({V}_{c}), we can choose a TF or motif (in our case, GATA) and ask which genes will be mostly affected by this TF by identifying the largest entries in the motif column. We used the default g_SCS multiple hypothesis testing correction to perform gene enrichment analysis and chose the top 1,000 genes. We filtered the results using term size (gene number in a term definition) greater than 500 and less than 1,000. Terms with adjusted P value less than 0.05 were retained as significant terms. The expression log10(TPM > 1) is used for the visualization against the GATA score.
In order to understand the relationships between the expression of targets and the expression of their target genes, we used this analysis. Gene-by-motif files were aggregated and organized into a unified structure comprising genes, motifs and corresponding cell features. We identified the target genes for each TF within predefined motif clusters and computed the mean expression levels of both the target genes and the corresponding TFs. To avoid potential artifacts in the expression measurement caused by experimental batch effects, we analysed both adult and fetal cell types, and fetal cell types only, and found similar results. The analysis was performed iteratively for all TFs within the motif clusters specific to fetal cell types.
The regulatory context for genes is embedded within a shared high-dimensional space using a cross-cell-type architecture. We collected the embedded genes from each transformer block of GET. The promoter’s output is defined as the embedding of a gene g. The embedding contains both promoter information and information from surrounding regions owing to the attention mechanism. The more space is dominated by the expression output, the deeper it is. tsne-cuda was used to visualize the embedding owing to data size. The clustering was done on the embedded space. The resolution is to keep the cluster number close to the UMAP density. For visualization, UMAP68 was used instead of cell-type-based subsampling.
We performed pairwise Spearman correlation in both cell-type specific and cell-type-agnostic settings. A matrix was constructed using input gradient scores. The correlation calculation was made using the genes with promoter overlap with open peaks in each cell type. Causal discovery was performed on the gene-by-motif matrix using LiNGAM69. For the cell-type-agnostic settings, 50,000 genes were randomly sampled from all cell types, and the resulting matrix was subjected to the LiNGAM algorithm implemented in the Causal Discovery Toolbox Python package with default parameters.
To be classified as an nested binder, we assumed that at least one of the two members of the family had a known interaction with a heterodimer in the STRING database. We thus used AlphaFold to predict the hypothetical homodimer structures of all known TFs and tried to predict whether a TF could be an intrafamily binder using AlphaFold-based metrics. We used several different AlphaFold-based metrics, including mean_plddt (average pLDDT score across all residues), pAE (predicted aligned error across all interchain interactions), pDockQ (predicted DockQ metric using interface pLDDT) and pDockQ × pAE. We found that pDockQ × pAE led to the best area under the receiver operating characteristic curve (0.69) and area under the precision–recall curve (0.41) when classifying intrafamily binder TFs.
pLDDT from AlphaFold is a reliable protein domain caller owing to its accurate structure prediction performance. We segmented each TF protein sequence into low and high pLDDT regions. Empirically, we found that 80% (recall) of known DNA-binding domains could be easily identified using high pLDDT regions plus a high ratio of positively charged residues. More specifically, we first computed smoothed pLDDT using a ten-amino-acid moving-average kernel and then normalized the score by dividing by the maximum. Any region that had a smoothed pLDDT score between 0.6 and 0.6 was defined as a low pLDDT region. If two low pLDDT regions were close (less than 30 amino acids), they were merged into one. Any region that was not a low pLDDT region was labelled as a high pLDDT region.
If the structure had a peak, we looked at it as evidence of interaction. Further checks to assess the confidence of the interaction were done. After Alpha Fold3 was released, we re performed structure prediction for a full length sequence and found the same PAX5 G182–NR domain interaction.
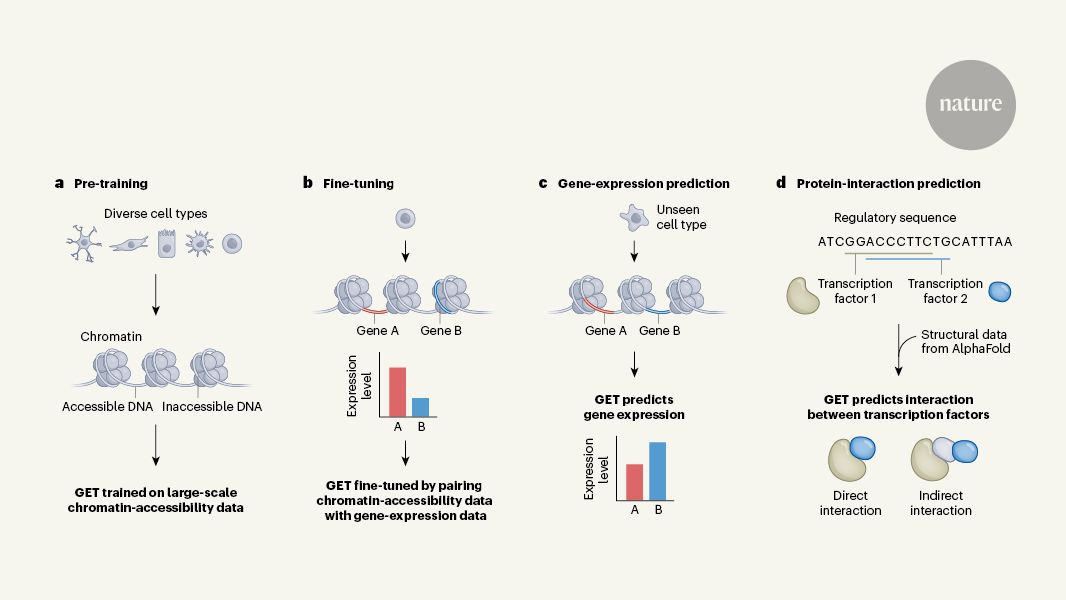
Source: A foundation model of transcription across human cell types
Transduction of ReH B-ALL HeLa cells into pCDNA3.1-MCS-13XLinker-BioID2-HA (Addgene, catalogue no. 11965)
The cells were purchased from ATCC. The cell lines purchased from the bank were not further authenticated. The cells tested negative for mycoplasma. There were no misidentified cell lines used in the study.
HeLa cells were cultured in DMEM (Gibco, catalogue no. 11965) supplemented with 10% defined fetal bovine serum (HyClone, SH30070), at 37 °C and 5% CO2. HeLa cell protein lysates were generated with 0.5% NP-40 lysis buffer (50 mM Tris-HCl, 150 mM NaCl, 0.5% NP-40) with a phosphatase and protease inhibitor cocktail (Sigma-Aldrich, PPC1010). Samples were incubated with 5 µg agarose-conjugated TFAP2A primary antibody (Santa Cruz Biotechnology, sc-12726 AC) overnight at 4 °C before being run in Laemmli loading buffer (BioRad, 1610737). After separation from the Tris–glycine gel, the proteins were transferred to the vacumm and probed with the primary antibodies against TFAP2A. The negative controls were probed with their primary antibodies against the SRF and -actin, and followed by the detection of chemiluminescence.
We initially cloned PAX5-WT and the PAX5 G183S mutant into the pCDNA3.1-MCS-13Xlinker-BioID2-HA (Addgene, catalogue no. There are 2 words in this sentence. We cloned the PAX5-WT-13Xlinker-Bio ID2-HA and the PAX5-G183S-13Xlinker-BioID2HA into a pCDH- GFP-puro vector. We transduced the REH B-ALL cell line (ATCC, CRL-8286) with pCDH-PAX5-WT-13Xlinker-BioID2-HA-GFP and with pCDH-PAX5-G183S-13Xlinker-BioID2-HA-GFP and selected transduced cells with puromycin (1 μg ml−1) to generate stable cell lines. The proximity labelling assay was performed following previously published methods71,72,73. There is a stable cell lines with control Vector pCDH-13Xlinker-HA-GFP. We collected the cells, washed them twice in cold phosphate-buffered saline and incubated them for 50 min on ice with occasional vortexing in lysis buffer (150 mM 10 mM Tris-Hcl, 10 mMK, 1.5 mM. Some of the protease and phosphatase inhibitors that were supplemented with are Life Technologies catalogue no. 78443 and sigma-Aldrich catalogue no. 70743-3). Cell separation was done at 21,000g for 15 min at 4 C. We performed total protein quantification using a Pierce BCA Protein Assay kit (ThermoFisher Scientific, catalogue no. 23225) and incubated 1 mg of total protein extract with 100 μl of magnetic streptavidin beads (Dynabeads MyOne Streptavidin C1, Life Technologies, catalogue no. 65002) on a rotator at 4 °C overnight to isolate biotinylated proteins. We washed the beads twice with lysis buffer, once with 1 M KCl, once with 0.1 M Na2CO3, once with 2 M urea 10 mM Twice, with the lysis buffer, Tris-HCL was pH 8.0. Biotinylated proteins were eluted by boiling in 4× protein loading buffer supplemented with 2 mM biotin and 50 mM dithiothreitol at 95 °C for 10 min. Biotinylated proteins in total protein extracts or immunoprecipitates were detected by western blotting using standard protocols and the following antibodies: streptavidin–HRP antibody (Life Technologies, catalogue no. Anti-NA, anti-HA, and anti-NRP2 are all included in the Cell SignalING catalogue. Proteins were detected using a Li-Cor Odyssey OFC instrument and quantified using the GelAnalyzer 23.1 software.
The experimental procedure involves the creation of a library of lentiviruses that contain both sequence elements and a mini promoter. The vector is randomly inserted into the genome through viral infection; the regulatory activity is then measured through sequencing and counting the log copy number of transcribed RNAs and integrated DNA copies.
Source: A foundation model of transcription across human cell types
Predicting Cell Type-Specific Three-Dimensional Contacts: From Enformer to GET via DistanceContactMap
This model can be improved if more input and learning are taken to predict cell type-specific three-dimensional contacts.
All genes were normalized across their 100 peaks to make them comparable across genes in this benchmark.
Recent studies have shown how important the one-dimensional genetic distance is in the governing of knockout effects of CRISPRi. In this benchmark, most methods include a component of genomic distance. For example, Enformer incorporates exponential decay in its positional encodings. benchmarking results follow the decay of the TSS after the DNA sequence has been incorporated into hyenaDNA. We’ve added distance information and extended the GET. In particular, we designed a simple DistanceContactMap module for GET to convert the pairwise one-dimensional distance map between peaks to a pseudo-Hi-C contact map. The distanceContactMap is a simple two-dimensional neural network of three layers with input and SCALE-normalized observed contact frequencies. The loss was used to train the model. We trained DistanceContactMap with the same K562 Hi-C data (ENCFF621AIY) used for training ABC Powerlaw, resulting in a 0.855 Pearson correlation, which mostly captured the exponential decay in contact frequency. The prediction of the model was termed “GET Powerlaw”. The other two scores shown in Fig. 3d are defined as follows:
HyenaDNA: we used the largest pretrained model available through Hugging Face (context length: 1 Mbp). To score the enhancer–gene pairs, we had to knock down the enhancer element and compare against the wild-type likelihood of observing the promoter sequence.
We used Enformer’s contribution score to make background normalization after Gschwind et al.33 described it.
Source: A foundation model of transcription across human cell types
Adapting the ABC Powerlaw to a broader population of cell types: Benchmarking and validation of the GET model for fetal cell types
The ABC Powerlaw was computed using the function in the official ABC repo with and the values that were trained on the Hi-C data in the same repo.
In our analysis and regulatory interpretation, we primarily used the binary ATAC model. The approach offers a way for the model to be attributed to sequence features, not just accessibility signal strength.
For example, we tested a ‘one-shot’ fine-tuning procedure using a single patient sample from a new dataset of patients with GBM. We compared the model against a pretrained model on 16 held-out patient samples. To ensure robust evaluation, we excluded two patients from this analysis to serve as a separate test set for assessing fine-tuning stability. The results were promising: fine-tuning on a single tumour patient sample enabled GET to achieve a Pearson correlation exceeding 0.9 when predicting expression for held-out patients, whereas zero-shot performance reached a 0.67 Pearson correlation. This demonstrates the model’s strong generalization capabilities and its potential for rapid adaptation to new datasets with minimal further training. As the availability of ATAC-seq and multiome data continues to grow, more comprehensive reference peak sets, such as the ENCODE DHS index13 and cPeaks65, will further facilitate the adaptation of the GET model to an even broader range of cell types and experimental conditions.
We validated the cross-cell-type prediction performance beyond astrocytes to include a broader range of cell types. The benchmark was performed on fetal cell types with a variable length peak set defined in the original fetal accessibility atlas1. This comparison features a quantitative ATACGET and binarized ATACGET as well as linear probing of Enformer CAGE output tracks, inferred genes in the Basenji test set, and a training cell type mean expression baseline. Pearson correlation, Spearman correlation and R2 were used to evaluate prediction performance.
Cell type rarity and library size: rare cell types often have smaller data libraries, which can limit the model’s learning potential and affect the accuracy of predictions.
Source: A foundation model of transcription across human cell types
RNA-seq prediction for the chromosome11 and astrocytes leaving out a K562-CAGE adaptor using Support vector Regression
We used scikit-learn Support vector Regression to do it. MultiOutputRegressor was used to handle two-dimensional output.
We have implemented comparisons with the following methods on the task of expression prediction when leaving out chromosome 11 and leaving out astrocytes, using the same input data as GET. The following parameters are used in our implementation.
QATAC from BATAC pretrain: in this setting, the base model was trained on the fetal–adult atlas with binarized ATAC signal; in the fine-tuning, we used the original aCPM ATAC signal.
QATAC from QATAC fine-tuned: in this setting, the base model was the leave-out-astrocyte RNA-seq prediction model trained on the fetal accessibility and expression atlas. This model was improved with the use of a quantitive ATAC signal.
These experiments leveraged LoRA parameter-efficient fine-tuning to achieve significant gains in time and storage complexity. All fine tuning coalesced in 30 minutes, making a K562-CAGE specific adaptor that could be merged into the base model.
Source: A foundation model of transcription across human cell types
ATAC prediction performance of a single cell type dataset using scATAC-seq data from EnCODE (accession: ENCFF998SLH)
To explore the impact of omitting motifs in the input features, we used K562 scATAC-seq data from ENCODE (accession: ENCFF998SLH) and evaluated the ATAC prediction performance when holding out randomly selected motifs. peaks with a threshold of q is the first we called. Then, we merged this peak set with the union peak set from the fetal pretraining data, keeping the peaks with at least ten counts in K562. For fine-tuning computational efficiency, we used LoRA parameter-efficient fine-tuning of the binary ATAC checkpoint pretrained on fetal and adult ATAC data with a 200-region receptive field (the pretrained checkpoint used for motif analysis in Fig. 4 and onward).
The performance was strong when they left out one to ten motifs. The performance was not good when using 20 motifs with a top 20% cutoff for each, because most of the training data had been removed.
It’s difficult to train a model on one dataset and apply it to another platform without proper tuning. Thus, for a new dataset with multiple cell types available, we took a leave-out cell type approach to fine-tuning. For a dataset of sorted cell types where only one cell type was available, we used leave-out chromosome training.
In summary, we used ATAC-seq and expression data from refs. 1,2,22. In total, the dataset encompassed 1.3 million single nuclei. The data was presented in a way that didn’t fit into standard formats. Cell types were from normal tissue. The pretraining dataset did not include any disease states. We incorporated further datasets in downstream tasks such as K562 and zero-shot analysis in tumour cells.
Source: A foundation model of transcription across human cell types
Learning to Forecast Regulative Elements in a Masked Autoencoder with Multi-Output Random Forest Regressor, Sckit-Learn and LoRA
Random forest uses scikit-learn Random Forest Regressor with max depth 10 and ten estimators. MultiOutputRegressor was used to handle two-dimensional output.
CNN: three Conv1d layers (layer dimensions: 283 input, 128, 64, 32, 3 kernel size) followed by FC(32, 512) → ReLU → FC(512, 2); SoftPlus was used for output activation. The same parameters were used in get and we didn’t change anything.
Low-rank adaptation can be used to perform fine-tuning over any layer. This is often used to adjust to a new platform, we apply loRA to the embedded layers and fine- tune the prediction head. This markedly reduces 99% of the parameters.
We used early stopping on the basis of validation loss to select the best models for later evaluation.
We adjusted the output for the regulatory-element-wise attention block. The layer normalization (LN), feed-forward network (FFN) and residual connections are finally used to generate the output for each layer. The mechanism behind the attention block can be summarized.
Similar to the Vision-Transformer-based Masked Autoencoders62, we replaced the regions in the selected positions with a shared but learnable ([{\rm{MASK}}]) token; the masked input regulatory element is denoted by ({X}^{\text{masked}}=(X,M,[{\rm{MASK}}])), where (X={{{x}{i}}}{i=1}^{n}) is the input sample with (n) regulatory elements. The training goal is to forecast the values of the masked elements. Specifically, we take masked regulatory element embeddings ({X}^{\text{masked}}) as input to GET, while a simple linear layer is appended as the prediction head. An overall goal can be formulated as:
The GET implementation is based on the PyTorch framework. We applied AdamW as our optimizer in the first training stage with a minimum weight decay of 0.05 and a max size of 258. The model was trained for 800 times and 40 times toggling for linear learning rate scaling. The learning rate is set at 1.5 104. The training usually takes around a week for a cluster with 16 V100 GPUs. AdamW63 was used for the second fine-tuning stage as an optimizer with a weight decay of 0.05 and a batch size of 256. The model is trained for 100 epochs and uses eight A 100 Graphics Processing Units. Inference for all genes in a single cell type takes several minutes, making it possible to perform large-scale screening.
We include a more detailed description of the optimization hyperparameters, computation infrastructure and convergence criteria used in the development of the model in the section below.
Computational infrastructure was similar to the pre training phase, and fine-tooling was done on eight A 100 Graphics Processing Units.
Epochs and duration: the fine-tuning process was shorter, consisting of 100 epochs, and completed in around one day. This phase was crucial for adapting the pretrained model to specific gene expression prediction tasks.
mz_l-1,;rmz_ +{{\rm{z}}}_{l}^{{\prime} },$$
Source: A foundation model of transcription across human cell types
Mapping Accessibility and Expression in Pseudobulks of a Fetal Cell Type Using Cell Barcodes
where ({W}{q},{W}{k}\in {{\mathbb{R}}}^{(n\times D)\times {d}{{\rm{k}}}},{W}{v}\in {{\mathbb{R}}}^{(n\times D)\times {d}_{{\rm{v}}}}) are learnable linear transformations.
The peak calling results from the original studies of the data were used to find a union set of peaks. To build a list of accessible regions, we excluded peaks with no counts.
In this study, the chromatin accessibility score for a specific genomic region was defined by the count of fragments located within that region for a given cell type pseudobulk. To enhance the generalizability of the model, these counts were further normalized through the logCPM procedure. Specifically, let t be the total fragment count in a pseudobulk, and let ci be the fragment count in region i. Then, the accessibility score si can be computed as:
The correspondence between accessibility and expression was determined through cell barcodes. Cell type annotations were used to facilitate the mapping when accessibility and expression were assessed on their own. Fetal expression atlas from Cao et al.23 was utilized for fetal cell types compared to adult data from tabula serenias 24 When several ATAC pseudobulk shared the same cell typeAnnotation, identical expression labels were assigned. This compromise was necessitated by the current dearth of multiome sequencing data, a situation expected to change dramatically in the near future.
Expression values were allocated to each region within our input. The values that were captured were not reflective of the current transcription rate due to the limitations of poly(A) scRNA-seq data. Valuable cell-type-specific information was provided by these values. The process begins by intersecting the input region list with GENCODE v.40 transcript annotation to pinpoint promoters, followed by assignment of logCPM values to regions corresponding to these promoters. All remaining regions are assigned a value of 0. The zero label on the non-promoter region of the cell helps to inform the model by showing transcription events happening in a cell.
The target input is a 200 2 matrix, symbolizing the expression levels of 200 regions across both positive and negative strands.