Researchers get an update on the Human Cell Atlas, and it is remarkable
A Census of Human Neural Organoid Cells and a Large-Scale Atlas of the Gastrointestinal Tract
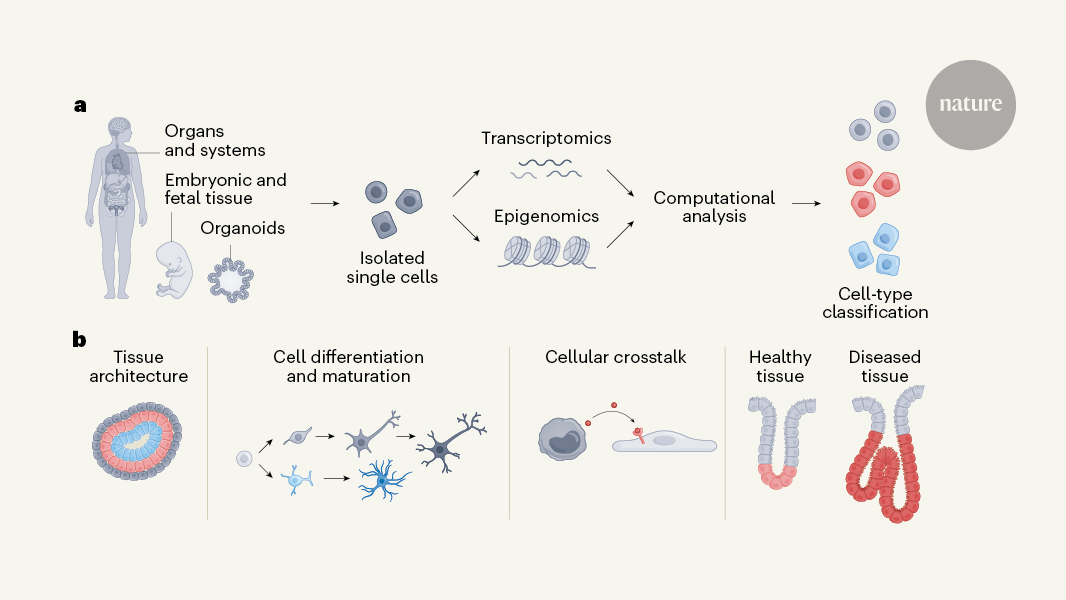
Why is this work needed? The building blocks of living things are cells. The Atlas can boost researchers’ knowledge of how the body works, while also being relevant to disease. Knowledge of where particular cells are located, combined with in-depth information about the genes they express and the epigenetic markers they carry, is revealing new insights into biology and disease.
The gastrointestinal tract atlas by Oliver et al.2 spans from the tissue of the mouth through to the oesophagus, the stomach, intestines and the colon. The current Atlas integrates previous data sets into a large-scale atlas of 1.1 million cells, with annotations of resident cell types and states. The data sets for the atlas include information about people with inflammatory diseases that affect the stomach, including coeliac disease and Crohn’s disease. Intestinal inflammation can cause cells to undergo metaplasia, a shift from one cell type to another. The authors used a mixture of data sets to compare the origin of metaplastic cells and stem cells. The benefit of the completeness of the atlas is that it can be used to compare the normal and abnormal states of cells in different organs.
Organoids have become a powerful model for functional analysis because of their intense brain study. The human neural organoid cell atlas is built on 1.7 million cells and involves 36 single-cell data sets and 26 protocols for producing organoids. The atlas is already shedding light on the crucial question of how faithfully organoids capture aspects of the developing brain. The authors found a correspondence between the length of time the organoids were in culture and the developmental stages that they resemble in the human brain: in the first three months of culture, the organoid resembles the cellular state of the fetal brain during the first trimester of pregnancy, whereas in the next three months, it resembles the second trimester. But the authors found an intriguing limit to this correspondence. The diversification in neuronal cell types that occurs with development did not continue in the organoid, and the fetal brain during the last trimester of pregnancy was not captured — leaving an open question about which required signals or other features are missing from organoid models.
Yayon et al.4 created a map of the thymus, a lymphoid organ that produces immune cells, in its early fetal development and early postnatal stages. Using the spatial dimension, the authors conceived of a ‘common coordinate framework’ to mathematically map the tissue. This model of the axis between the outer part of the thymus and its centre (the cortico-medullary axis) allows for a deeper understanding of tissue organization and comparison of the organ both in and between individuals. Old age is a stage of life that will be interesting to study.
There is a coordinated self-organization of continuously differentiating cells that takes place during fetal and embryonic development. Despite the immense complexity of these processes, scientists have a limited understanding of the cell mechanisms that lead to such events.
They explored the development of the skull and joints of the limbs for up to 1 week after conception. The identification of the key gene-regulatory networks that direct the commitment of the cells to chondrogenic and osteogenic lineages was accomplished through simultaneous mapping of transcriptomic and epigenomic profiles of single cells. The authors inferred probable lineage relationships along differentiation pathways and propose how cellular crosstalk might guide the formation of bone, identifying a potential key role for interactions with the vascular system. The authors combined the data from their single-cell analysis with the genome-wide association studies to identify developmental cell states that could be linked to osteoarthritis and other diseases in adult skeletons.
Similarly, Gopee and colleagues present a comprehensive cellular atlas of skin development spanning 7–17 weeks after conception. Using a combination of single-cell and spatial transcriptomic technologies, the authors mapped dynamic changes in cell states and detail how these cells organize to form developmental structures and interact in microanatomical skin niches. Their findings highlight the unexpectedly diverse role of immune cells in coordinating developmental processes, particularly the involvement of macrophages in the formation of blood vessels by endothelial cells. An innovative organoid system was used to recapitulate key aspects of skin development.
popV is a model for transferring cell-type labels from annotated to unannotated data sets. The popV model is an ensemble model, meaning it combines prediction from existing models with the level of disagreement between the underlying tools to produce both cell-type labels and uncertainty scores. This approach highlights ambiguous cases, which reduces manual review and draws attention to cell populations that are challenging to classify. This feature reduces the load on researchers, makes popV adaptable to future models, and improves the interpretability of results.
MultiDGD tackles integration of multiple data points, such as gene expression and the accessibility of chromatin, using a deep variable model. MultiDGD learns the optimal hidden-variable representations that are shared across all data sources without the need to define important features. By incorporating information about potentially confounding variables, such as inconsistencies between samples, multiDGD enables post-hoc data integration across data sets, making it suitable for multi-omics studies, in which data were gathered from different sources. The model improves alignment of data and allows associations of genes and regulatory regions of the genomes to be mapped in an essential step in understanding gene-regulatory networks.
Together, popV and scTab lead efforts in standardized annotation and consensus-building, whereas multiDGD opens up avenues for data integration across complex multimodal data sets.
This does not lessen the impact of these methods, but rather highlights the field’s rapid pace and the importance of innovation. To meet this growth, future research is likely to emphasize adaptable and interoperable solutions. These methods contribute valuable foundations for future advancements, paving the way for even more adaptable and scalable models for single-cell, multi-omics data.
The Global Human Cell Atlas (HCCA): Where do we stand? Where are we, where do we go? How do we look at the world, where are we going?
The studies from the global project are a major accomplishment, coming less than a decade after its launch. Funders should sign up for the long haul.
Findings from researchers working on the lung cell atlas, for example, highlight the differences between the lungs of a sample of people in Malawi who died from COVID-19 and those who died from other lung diseases2. Scientists have looked at the development of organs through analyses of human fetal skin and craniums.
The HCA would not have been possible without earlier projects, notably the Human Genome Project and, more recently, the NIH BRAIN Initiative, as well as ENCODE, a project to build a ‘parts list’ of functional elements in the human genome. The HCA teams have also worked hard to reflect human diversity in their data. Scientists from Africa, Asia, Latin America, and the Middle East are part of the consortium. Researchers from these regions were invited not only to join, but also to help lead and coordinate HCA projects, and to do so according to priorities relevant to local populations. The initiative now involves more than 3,000 scientists across 1,700 institutions, recording and studying data from people in around 100 countries.
Large-scale consortiums have a limited lifespan, as do most research projects. Ten years is considered generous. A handful of projects might last a few years longer. Permanent funding tends to be reserved for projects of national or international importance, including essential infrastructure — the tools and technologies without which vital discoveries and inventions would not be possible. That is how the HCA should be compared to.
In what cells are active in disease-associated variants of the human genome? A pedagogical report on the HCA Genetics Working Group
“While genetic studies have mapped more than 100,000 disease-associated variants in the human genome, we do not know in which cells the majority of these variants are active,” HCA researchers write6. They state that without this knowledge, we cannot fully understand biology, study more powerful models of disease, deploy better diagnostics and develop more effective therapies.