A big study shows that eating less can lead to a longer life
Generalized additive mixed models for assessing the correlations between diet, weight and age in a diverse population: A case study of 960 mice with a genetically distinct population
The researchers monitored the health and lifespan of 960 mice, each a genetically distinct individual from a diverse population, that mirrored the genetic variability found in humans. Some mice were on a diet of very little calories while others are allowed to eat freely.
Outside investigators look at the results to see the intricate and Individualized nature of the body reaction to caloric restriction. James Nelson is a biogerontologist at the University of Texas Health Science Center in San Antonio.
The study was published in Nature by the author and his co-authors, as well as scientists at the anti-bacterial company that funded the study.
Daniel Belsky cautions against over-politating from mice to humans while he is an epidemiologist at Columbia University. He acknowledges that the study adds to the growing belief that lifespan and healthspan are not the same thing.
For traits collected annually or biannually, we were able to explore hypothesized direct and indirect relationships with diet, body weight and age. We used generalized additive mixed models (GAMMs) with a gaussian/identity link to analyse these effects by fitting a series of nonlinear relationships between trait response and covariates. GAMMs for a combination of fixed and random effects (formulae below) on trait response pre-adjusted for batch were fit using the gam() function of the mgcv package in R, with the Newton optimizer and default control parameters. The percentage of life lived was rescaled to reflect the age at the test. The PLL scale removes artifacts due to survivorship bias across groups with different lifespans. Before model fitting all continuous variables were rank normal scores transformed.
The reduced representation of23 features was recomputing a sparse low-rank network on which we obtained a covariance decomposition. The relative importance of different effects of DR on lifespan is calculated using the reduced representation graphical model. The absolute values of path scores were used to rank the relative importance of each path and normalized to sum to 1 to estimate the fraction of the covariance between DR and lifespan explained. To obtain network visualizations, the position of nodes and orientation of edges were determined by computing max-flow through the path network. We define the path network as the graph formed by taking the partial correlation network but reweighting edges to be the sum of all (absolute) path scores of paths that contain that edge.
where T is trait, D is dietary assignment, BW is body weight at the date closest to T’s collection date, PLL is the proportion of life lived as of T collection date and s() is the smoothing parameter. Each mouse had multiple datapoints across the T collection date. This clustering was accounted for with a random intercept for ID, specified as (1|ID) above. We performed hypothesis tests to find out trait sensitivity to body weight, and diet-by-trait interaction. We identified traits that responded to body weight in a way that was consistent with the models specified above, as well as the ones that responded in a similar way to diet. Health, metabolism, haematology and immune were categorized. For each trait category, bar plots were generated to show the number of traits with significant (FDR < 0.01) associations with body weight, diet, PLL and diet × PLL interactions. The same analysis was done with age in months. We applied FDR adjustment to each test across traits and timepoints (Benjamini–Hochberg FDR method).
Where BWtest is at the time of testing, and where BW6 is the last preintervention body weight. Body weight terms were not used for the body composition and change-in-body weight traits. All continuous variables were ranked normal scores before fitting a model. The test we performed was for the diet and body weight adjusted association. We applied a FDR adjustment to each test across traits and timepoints (one-step Benjamini–Hochberg method). Traits were categorized and significant (FDR < 0.01) results were tabulated as above.
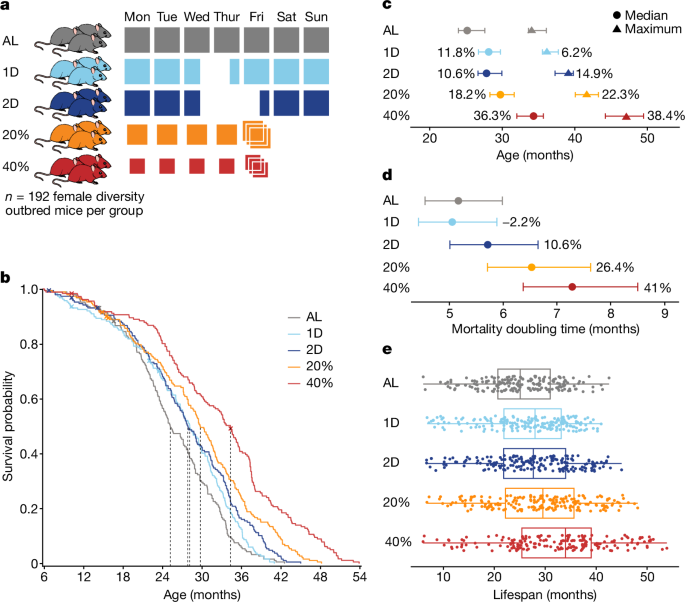
Climber was used to track animals, schedule testing and provide stable repository for primary data collection. The data was regularly reviewed by a statistical analyst. Initial data quality control included identifying and resolving equipment miscalibration, mislabelled animals and technically impossible values. If we could not manually correct these using laboratory records, they were removed. Quantitative assays including body weight and temperature were explored for outliers. Quantitative traits other than body weights were corrected. To quantify batch effects, we fit a fully random-effects linear mixed model conditioning on diet, body weight at test and age. We adjusted trait values by subtracting the batch model coefficients. Lifespan data is presented in months for easy interpretation. In the main text there is a report of P 2.2 1016 and non-truncated P values are provided in the Supplementary Information. The analyses were done using R v.4.2.2 and RStudio. There are data analysis scripts available.
Research staff regularly evaluated mice for prespecified clinical symptomology: palpable hypothermia, responsiveness to stimuli, ability to eat or drink, dermatitis, tumours, abdominal distention, mobility, eye conditions (such as corneal ulcers), malocclusion, trauma and wounds of aggression. If mice met the criteria for observation in any of these categories, veterinary staff were contacted. If the clinical team determined a mouse to be palpably hypothermic and unresponsive, unable to eat or drink, and/or met protocol criteria for severe dermatitis, tumours and/or fight wounds, pre-emptive euthanasia was performed to prevent suffering; otherwise, the veterinary staff provided treatment. The survival curves show deaths in the mice euthanized or found dead. The mice euthanized due to unrelated injuries to imminent death were treated as censors.
Free wheel-running data was collected for three and a half years. Mice were individually housed for a minimum of 36 h in a special cage suited to house the Med Associate low profile running wheel with a wireless transmitter. The food hopper was removed to make it easier to move the wheel and put food on the floor. The electronic base tracks the revolutions as the wheel sits at an angle. The battery-powered base allows continuous monitoring of data, which is transmitted in 30 s intervals to a local computer.
A piece of blotting paper was cut to fit standard duplex cage dimensions. Shavings were removed from a clean cage, and the paper was taped to the bottom of the cage. Food was provided during this test; however, water was removed to prevent possible leaking onto the blotting paper. Four Mice were individually housed in a cage. The mice were returned to their previous housing units at the end of the trial, while the papers were taken away and dried for up to 4 hours. The papers were taken to Beth Israel Deaconess Medical Center, where they were scanned with the ultraviolet light in order to quantify the void spots.
We used the Ugo–Basile rotarod, which has five lanes evenly spaced along a motorized horizontal rotating rod, allowing for up to five mice to be tested simultaneously. A platform beneath each lane is used to record thelatency for each mouse to fall. At the beginning of the session, mice were placed onto the rod, which began rotating at 4 rpm, slowly increasing to a maximum of 40 rpm, over 300 s. Mice were given three consecutive trials. We reported the mean latency (time to fall) and the slope of latencies across trials, as well as the number of trials with no falls and number of trials with immediate falls. In case a mouse didn’t cooperate, trials were recorded as missing.
Startle response was measured in rodents using an automated startle chambers in which a mouse is placed in a clear tube attached to a platform that is sensitive and sensitive to noise while being exposed to various stimuli at varying decibels. A number of randomized, computer-generated stimuli ranged in volume from 70 to 120 decibels at 40 ms and an interval of 9–21 s, which were triggered by white noise from the overhead speaker. The test runs for approximately 30 min.
Ultrasonography of NK and T Cells in Mouse Bocytes with Fasted and Non-fasted Glucose Measured through Pulsed Doppler Sonography
In order to do ultrasonography, 30 and 40 MHz probes were used. Echocardiography uses pulsed Doppler sonography, applied through the ultrasound probe, to measure blood flow rates and volumes.
At the flow cytometry blood collections at 16, 62 and 114 weeks, mice were fasted for 4 h and glucose was measured using the OneTouch Ultra2 glucose meter from LifeScan along with OneTouch Ultra test strips. Non-fastedglucose was measured at each of the three blood collections.
It was possible to identify subsets of T cells that were naive because of outbred nature of these mice and the presence of CD62L and CD44. NKG2D-positive cells were enumerated and may represent memory T cells that accumulate after immune responses60. The outbred DO mouse line has limitations in the identification of NK cells and their subsets, which is why CD11c and CD11b have been used within this population. CD11c is reduced on the least mature subset of NK cells.
Flow cytometry can be used to determine the frequencies of circulating immune cell subsets. Analysis was performed before the start of dietary interventions at 5 months, then at 16 and 24 months of age. These timepoints corresponded to 11 and 19 months of dietary intervention. Red blood cells were washed and lysed before being put into a buffer. Cells were resuspended in 25 l FACS buffer. Antibodies including the Fc block were added and prepared for 30 min at 4 C. Labelled cells were washed and DAPI was added before analysis on the LSRII (BD Bioscience) system. The antibody cocktail contained CD11c FITC, N418 (35-0114-U100, Tonbo Biosciences, 1:100); NKG2D (CD314) PE, CX5 (558403, BD Biosciences, 1:80); CD3e PE-CF594, 145-2C11 (562286, BD Biosciences, 1:40); CD19 BB700, 1D3 (566411, BD Biosciences, 1:40); CD62L PE-Cy7, MEL-14, (60-0621-U100, Tonbo Biosciences, 1:100); CD25 APC, PC61 (102012, BioLegend, 1:80); CD44 APC-Cy7, IM7 (25-0441-U100, Tonbo Biosciences, 1:40); Ly6G BV421, 1A8, (562737, BD Biosciences, 1:80); CD4 BV570, RM4-5 (100542, BioLegend, 1:40); CD11b BV650, M1/70 (563402, BD Biosciences, 1:160); CD45R/B220 BUV496 (RA3-6B2, 564662, BD Biosciences, 1:20); Fc Block, 2.4G2 (C247, Leinco Technologies, 1:100).
Source: Dietary restriction impacts health and lifespan of genetically diverse mice
Multi-dose dual X-ray absorptiometry with the LUNAR PIXImus II densitometer
We performed dual X-ray absorptiometry analysis using the LUNAR PIXImus II densitometer to collect bone density and body composition (including fat and non-fat lean tissue). There were disposable plastic trays that the mice were put on before being placed onto the exposure platform. The process takes approximately 4 minutes. Measurements were taken at around 44, 96 and 144 weeks of age.
The modified version of the clinically relevant FI38 was put to use as an average of 31 signs of age-related deficits and health decline. Each trait was scored on a scale of 0, 1 or 5, where 0 indicated the absence of the deficit; 0.5 indicated mild deficit; and 1 indicated severe deficit. The baseline measurement was taken for 5 months, and then repeated every 6 months. Simple averaging yielded a raw FI score of between 0 and 1 for each mouse. Frailty scores were adjusted by estimating batch, coat colour and experimenter effects as random factors that were subtracted from raw frailty score values before statistical analysis.
Over 100,000 values were collected over the course of their lives from the 937 mice. The weights were analysed after the local regression fitting within the mouse.
To obtain an accurate estimate of food intake and changes in body weight in response to weekly fasting cycles, we set up an independent cohort of 160 female DO mice. Mice were placed on the same DR protocols as in the main study. Food was weighed daily for a period of 1 week when mice were 30, 36 and 43 weeks of age. The food intake data was normalized and presented as daily and weekly averages for timepoints by diet. At 43 and 45 weeks of age, the body composition was determined using non-imaging nuclear magnetic resonance (NMR), a device with a 5-gauss magnet that is adapted to small- animal studies. Before and after fast, the data was used to detect changes in body weight and composition. There were values of body weight, fat mass and adiposity that were co-plotted with the difference between before and after fasting.
DR was implemented by controlling the amount and timing of food given to mice. Feeding schedules for DR were started at 6 months of age. All mice were fed a chow diet. The AL feeding group was provided with unlimited access to food and water. The IF mice were provided unlimited access to food and water. On Wednesday of each week at 15:00, IF mice were placed into clean cages and food was withheld for the next 24 or 48 h for the 1D and 2D groups, respectively. CR mice were provided with unlimited access to water and measured amounts of food daily at around 15:00, 2.75 g per mouse per day for 20% CR and 2.06 g per mouse per day for 40% CR. We estimated the amounts using historical feeding data from DO mice, which showed an average daily consumption of 3.43 g per mouse. The mice were subjected to 20% CR for 2 weeks, then 30% CR for an additional 2 weeks, and finally 40% CR for the entire protocol. In the 2D IF protocol, mice were initially acclimatized to the 1D IF regimen for 2 weeks. Mice were co-housed with up to eight mice per pen. Co-housing is standard practice for CR studies39; competition for food was minimized by placing food directly into the bottom of the cage, allowing individual mice to ‘grab’ a pellet and isolate while they eat. CR mice were provided with a 3 day ration of food on Friday afternoon, which resulted in weekly periods of feasting followed by a period of food deprivation of approximately 1 day for the 20% CR mice and 2 days for the 40% CR mice, comparable to the IF fasting periods. The feeding time is close to the normal time of day for mice, starting just before the start of the dark cycle and lasting until the end of the day. The lifespan extension of mice subjected to 30% CR8 was maximized by this timing. The feast–famine cycle can be used to assess the health impacts of CR57,58,59 but there is no direct assessment.